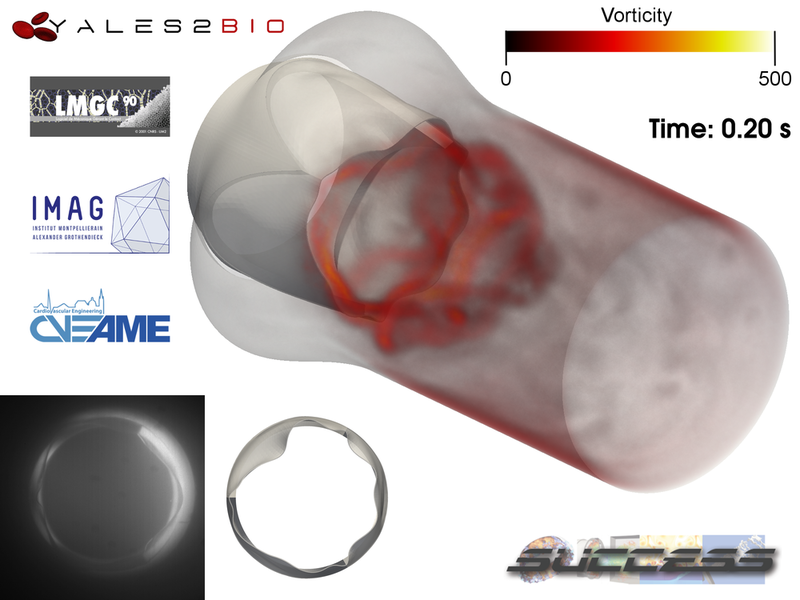
Flow through a model of aortic valve
We present here a 3D unsteady FSI computation of the hemodynamics through a model of an aortic valve, computed by J. Siguenza thanks to YALES2BIO. The fluid-structure interaction problem is solved by coupling YALES2BIO and LMGC90, the latter solving the structure problem. The objective is here to combine 3D FSI simulations, with experimental stereoscopic Particle Image Velocimetry (PIV) measurements, to validate the leaflet kinematics and the blood flow description. On the bottom left corner of the video, experimental and computed leaflet kinematics are compared, showing an excellent agreement.
This combined experimental and numerical study was performed in a joint project between IMAG and the Department of Cardiovascular Engineering of the Helmholtz Institute, in Aachen. This is a collaboration with Desiree Pott, Simon Mendez, Simon Sonntag, Ulrich Steinseifer and Franck Nicoud. This work has been supported by the yESAO Exchange Program, of the European Society for Artificial Organs.
The coupling between YALES2BIO and LMGC90 was achieved in a joint project between IMAG and LMGC, the laboratories of Mathematics and Mechanics of the University of Montpellier, respectively. This is a collaboration with D. Ambard, F. Dubois, F. Jourdan, S. Mendez, R. Mozul, F. Nicoud.
More details can be found in Siguenza et al., IJNMBE, 34(4), 2018. See also Siguenza et al., JCP, 322, 2016 for more details about the numerical method and benchmark test cases.